Code
You can find below the code developed and data used in my research projects.
Analysis of geometrical properties of 3D meshes
Scripts implemented in python to quantify geometrical properties of 3D meshes using Blender software. They have been used in Denizot et al., Glia, 2025. The code is available on Github and meshes on Zenodo.
Generation of realistic perisynaptic astrocytic processes meshes with various endoplasmic reticulum distributions and constant shape
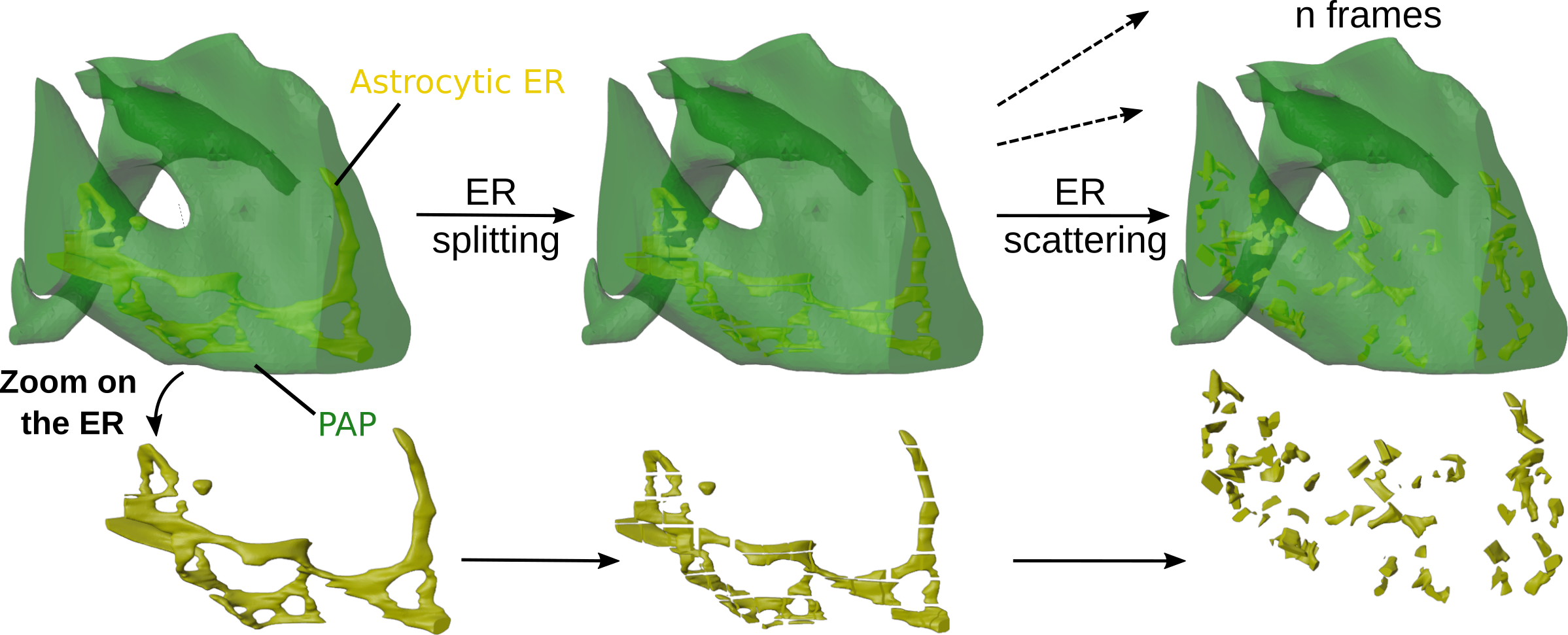
Scripts implemented in python to generate 3D meshes of perisynaptic astrocytic processes with various ER distributions while maintaining constant volume and shape using the Blender software. The code and resulting meshes are available on Github and were used in Denizot et al., Glia, 2025.
Simulations in idealistic 3D geometries of fine processes based on recent super-resolution microscopy data
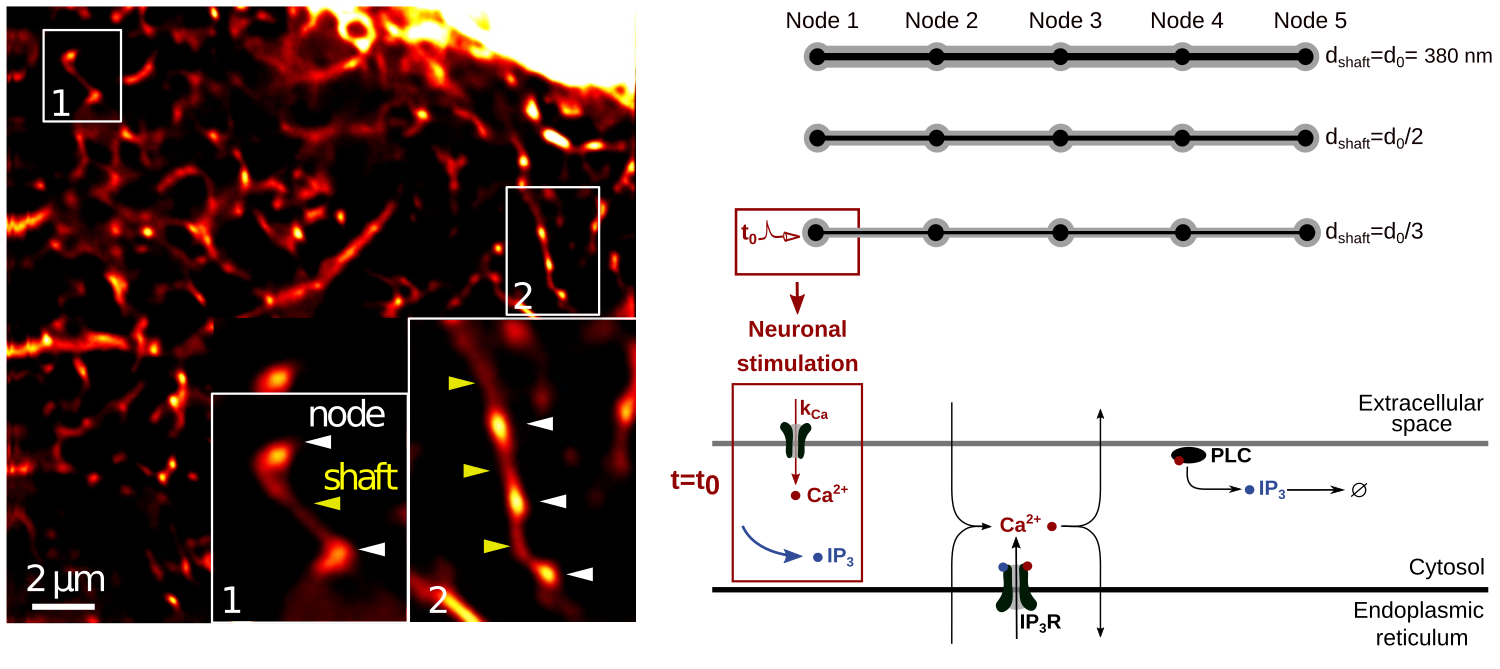
Simulations published in Denizot et al., bioRxiv, 2021. Reaction-diffusion simulations were performed in 3D meshes that were designed from the latest data available from super-resolution microscopy on astrocytes from Arizono et al., Nature Communications, 2020. The high spatial resolution of this model allows to propose plausible mechanisms by which astrocyte morphology at the nanoscale, notably shaft width, can influence local calcium dynamics and thus affect specialized neuron-astrocyte communication. The simulation code and 3D meshes designed in this study can be downloaded here.
# IP3R-dependant Ca2+ signaling in fine astrocytic processes 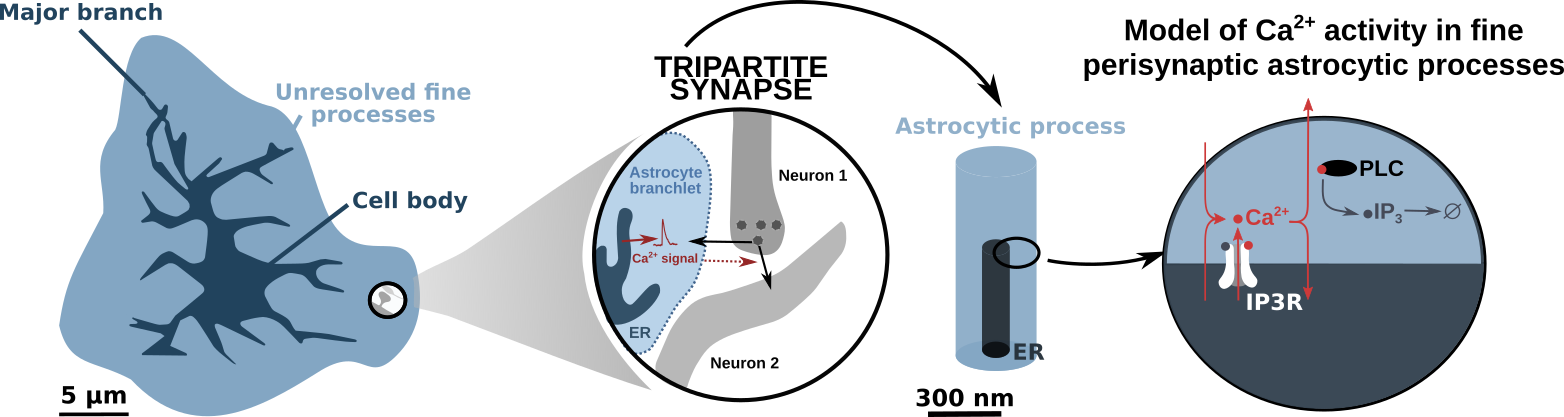
This model has been developed and presented in Denizot et al., PLOS Computational Biology, 2019. The code can be found here. 4 different implementations of the model are available:
- ODEs: deterministic, well-mixed implementation of the model with XPPAUT software
- Gillespie: stochastic, well-mixed implementation of the model in Python
- Particle-based: stochastic, spatial, in 2 spatial dimensions, implemented in C language
- Voxel-based: stochastic, spatial, in 3 spatial dimensions, implented in python, using STEPS software
